By Marta Koblańska, January 29, 2026, 16:00 Poland’s time. The Author of the photo: Ruijin Ji, Cell Reports
By modifying the conversion of purine bases constituting a body template for growth and health, there could be possible to cure the most damaging illnesses, according to the scientists in China.
China is well-known for its farsighted medical experiments. The country also has more flexible legal rules to conduct them, while it has switched toward conditions for experiments from ex vivo to in vivo. The recent outcomes have shown awesome achievement. In the new study published on January 20, 2026, in the journal ,,Cell Reports Medicine,” scientists present a new approach, thus based on previous discoveries of the CRISPR approach to the naturally occurring process of DNA and RNA repairing due to random or facilitated mistakes/errors, which may lead to various deficits or health problems in eukaryotes.
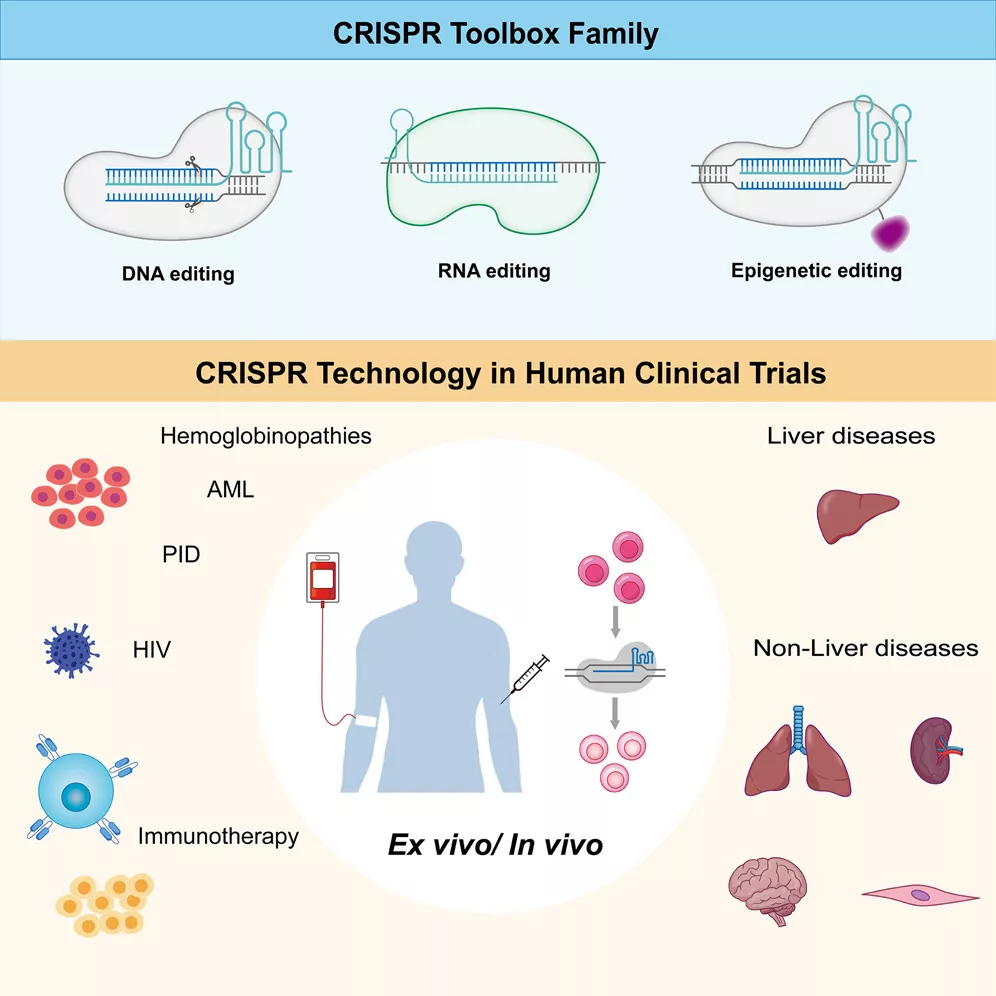
The major achievement is thus that using biological/genetic engineering, we can change the location or conversion of purine bases in a given section of our DNA or RNA, with an emphasis on the single-stranded nucleic acid RNA. And this is absolutely newest, as previously, the CRISPR tool, technologically enhanced, was first of all dedicated to cutting off the damaged fragments of the genome, thus enabling elimination of fatal or health-destroying mistakes.
These purine bases, being a rather stable template of us, build up the deoxyribonucleic acid DNA and ribonucleic acid RNA. There are four purine bases for each template, which bind with each other or other compounds, as well as convert in a certain way. The incredibility of the Chinese experiment is based on the repair potential during an editing window, when an insertion is performed by infusion of curing nuclease/deaminase solution to the harmed/degraded location in the genome.
What is absolutely overwhelming?
The DNA or RNA templates are not stiff building blocks, but rather act as dynamic transforming substrates; the damage in the process of transformation or the degradation of the base acids can be reversed, for now, in laboratory conditions. The template accepts the donated infusion and undertakes the task of further transformation in line with the newly passed instruction. At the same time, the modified location is avoiding,, error-prone repaired pathways”, as scientists write.
Why is this experiment/study so important?
Some may say it represents a strong interference into the chemical and biological processes, which enable the constituting future cell proliferation and growth by grasping the primordial single-strand RNA base, yet during a specified phase of its transformation. The truth is also that RNA errors can later lead to breaks in the double-stranded DNA. The authors of the study say that the editing of the template base via insertions or deletions of a chemically isolated and biologically active solution/base (MK’s note) without cutting both DNA strands in the body, which is aimed to be edited (MK’s note), offers potential therapy, when permanent DNA changes are undesirable.
Beyond nucleases that induce double-stranded breaks (DSBs), platforms such as base and prime editors enable precise nucleotide substitutions, insertions, or deletions without cutting both DNA strands. Base editors enable single-nucleotide changes without generating DSBs or requiring donor DNA templates, write Ruijin Ji, Quibing Chen, Ying Zhang of Zhongnan Hospital, TaiKang Centre for Life and Medical Sciences, and State Key Laboratory of Virology and Biosafety, University of Wuhan, China.
The key to the study represents nucleases and adenosine deaminases, acting on RNA (ADAR) enzymes, which enable transient RNA base conversions. In short words, you add a block/molecule in the place that originally has been damaged, or you remove a block/molecule that inhibits the endogenous repair pathway of DNA. And you can take it from a prehistoric organism – a virus which is built only from a single-stranded RNA and called an organism in transition between living and dead, or a bit more advanced – bacteria being recognised as living.
All of the engineering is possible under the condition that you can do it on time, i.e., in a certain phase of division, which ( a paradox) leads to the growth or conversion, during which the purine bases (specified either in structure or in pairs they bind to or cleave) can change.
How do the authors of the study describe their achievement?
– Two major classes of base editors have been developed. Cytosine base editors (CBEs) mediate C⋅G-to-T⋅A conversions by deaminating cytosine to uracil, creating a U⋅G mismatch that is resolved as a T⋅A pair by DNA polymerase. An uracil glycosylase inhibitor (UGI) is often included to prevent uracil excision and enhance editing efficiency. Adenine base editors (ABEs) convert A⋅T to G⋅C by using an evolved adenosine deaminase (derived from Escherichia coli TadA) to deaminate adenine into inosine, which is read as guanine by the DNA polymerase. Base editing occurs within a defined editing window of the single-stranded DNA (ssDNA), where the sgRNA hybridises with the target strand, displacing the non-target strand and exposing PAM-distal nucleotides as ssDNA.These nucleotides are accessible to the deaminase, and their compatibility with the catalytic domain determines the efficiency, position, and specificity – write Ruijin Ji, Quibing Chen, Ying Zhang, who equally contributed to the research.
– A major challenge in the clinical application of genome editing tools lies in the efficient and targeted delivery of CRISPR tools to specific cells or tissues. Effective delivery systems must ensure high intracellular uptake while minimising cytotoxicity, immune activation, and off-target delivery. Current approaches can be broadly categorised into three classes: physicochemical methods, viral vectors, and non-viral platforms, each with distinct advantages and limitations, claim scientists.
The work was supported by the Chinese public institutions and organisations, as well as postdoctoral programmes.
Leave a Reply